The OpenData movement has the White House producing and releasing some novel datasets. One of them is the We The People petition site. I learned about this from Proofreader.com’s interesting python work using that data. From the petition site, you can see an interesting gallery of work done in different language and for different media/platform. One such example is yoni’s r pkg.
In the spirit of the open data goodness, I thought I’d play around as well with this d3 chord diagram.
Read on to make your own!
Downloading data from github repo
if (!require("pacman")) install.packages("pacman")
pacman::p_load_current_gh("mattflor/chorddiag")
pacman::p_load(dplyr, magrittr, ggplot2, tidyr, curl)
So proofreader’s site has a csv, but I found yoni’s pkg most useful because it had scraped the API for more data both in terms of rows and columns. If you wanna hit that API now, download yoni’s package. It’s supposed to have functions for connecting to the API, but I didn’t try it.
curl::curl_download(
"https://github.com/yoni/r_we_the_people/blob/master/data/petitions.RData?raw=true"
, destfile="~/Desktop/petitions.RData" )
load("~/Desktop/petitions.RData")
So I found myself that download.file was a bit glitchy (trying to download the github website instead of the raw data), so curl::curl_download was my go-to, probably will be from now on. Make sure to use the link to the raw data (as I did) in your curl call, using the ?raw=true at end of url.
Cleaning / Set up
There’s been some really nice work nice work in visualizing data with d3 and
exposing those tools through APIs for R. htmlwidgets has been at the fore of this. (Thanks y’all!)
A fun pkg I’ve recently found for making chord diagrams is Matt Flor’s chorddiag github pkg. So all my data cleaning work will be towards getting an adjacency matrix of tag counts.
# recover tag names and ids
p <- petitions # save some typing
ids_names <- rbind(
p[, c("issues1.id", "issues1.name")] %>% setNames(c("ids", "names"))
, p[, c("issues2.id", "issues2.name")] %>% setNames(c("ids", "names"))
, p[, c("issues3.id", "issues3.name")]%>% setNames(c("ids", "names"))
) %>%
unique() %>% na.omit()
# get only petitions with multi-tags
tag_count <- p %>%
select(id, issues1.id, issues2.id, issues3.id) %>%
tidyr::gather(order, cats, -id) %>%
filter(!is.na(cats)) %>%
mutate(order = tidyr::extract_numeric(order)) %>%
left_join(ids_names, by=c("cats"="ids"))
With these chord diagrams I think a healthy check with a more traditional visualization gives you some barrings on what to expect from the chorddiag output. It also gives you some ways you might want to tweak options, as will be demonstrated below.
xtab_tag <- tag_count %>%
count(names) %>%
arrange(desc(n))
xtab_tag %>%
ggplot2::ggplot(aes(x=factor(names,levels=names),y=n)) +
geom_bar(stat="identity") +
theme(axis.text.x = element_text(angle = 45, hjust = 1, size = 12),
plot.margin = unit(c(10,10,20,20), "mm"),
plot.title = element_text(size = 20)) +
ggtitle("Distribution of All Tags") +
labs(x = "petition categories",
y = "category counts")

Adjacency Matrix
Now we build the matrix by registering whether a tag shows up for a specific petition and then creating the adjacency matrix to represent co-occurences of tags, which is what we need for the chord diagram.
# list of tags
tags <- sort(unique(ids_names$names))
# matrix to hold counts
mat <- matrix(0,nrow=nrow(tag_count),ncol=length(tags))
colnames(mat) <- tags
# get columns with tags from dataframe
p_id_nam <- p %>%
select(contains(".name")) %>%
mutate(issues1.name= ifelse(is.na(issues1.name), issues.name, issues1.name)) %>%
mutate_each(funs(ifelse(is.na(.), "", .)), starts_with("issues"))
# make matrix
for (i in seq_along(tags)) {
for (j in c(1,2,3)){ # 1,2,3 are columns I want
mat[,i] <- as.numeric(tags[i]==p_id_nam[[j]]) + mat[,i]
is.na(mat[,i]) <- 0
}
}
adjmat <- t(mat) %*% mat
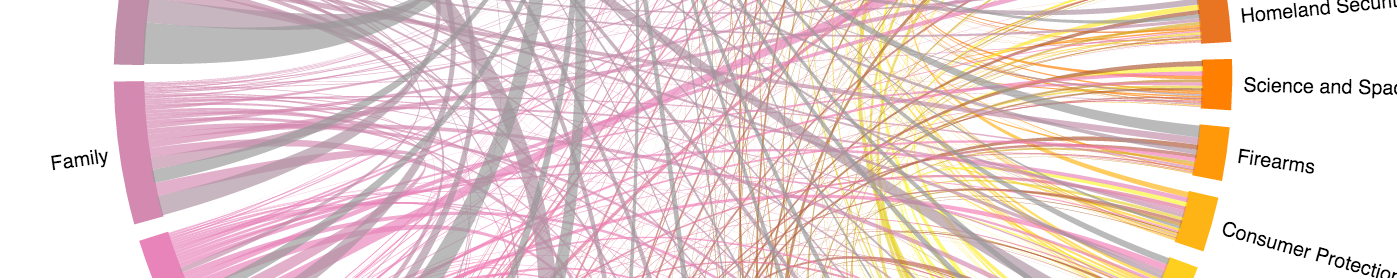
Chord Diagram and choices
I’ll save you the suspense regarding what choices I had to make in order to get
chorddiag to work for me.
The first is that this data set has many more categories or groups than chorddiag is set up to work with out of the box. This is mainly because they have a default color selection referencing RColorBrewer palettes, which have fewer than we need.
So this blogpost was really helpful in showing how you can augment RColorBrewer palettes for many
more than the usual set. It build a function that can then create as many colors as you need off
variations in an original ColorBrewer set. This function shows up below in the chorddiag groupColors
option where you pass in a palette other than the default.
# set number of colors needed
colorCount <- length(tags)
# makes function to create palette
getPalette <- grDevices::colorRampPalette(RColorBrewer::brewer.pal(9, "Set1"))
The second choice is that the order of how the chord are printed can really obscure
the main take away from your pre-attentive processing. Thus, I used a function to order the
adjacency matrix by the categories with the most. This ordering allows the smaller categories to show up
in the background, leaving the more prominent categories unobscured. One can also change the
chordedgeColor option to something fainter to lessen their influence further.
# manage use of diagonal cells in adj_mat
remove_diags <- function(mat, rm.lower = TRUE, ...) {
diag(mat) <- 0
if (isTRUE(rm.lower)) mat[lower.tri(mat)] <- 0
mat
}
# ## order plot layering by smallest to largest so larges are on top
ord <- order(rowSums(remove_diags(adjmat, FALSE)))
Third choice was whether you wanted the diagonal of the matrix to show up.
In the diagonal the counts represent the tags own association with itself. So visually the chord in the
diagram will return unto itself, forming a hump of sorts, for that category. The humps
work sort of like the bars in barchart above in showing the relative weight each category had on its own,
independent to how it is connected to other tags.
# with the diags means there's a return
chorddiag::chorddiag(adjmat[ord, ord], margin = 150, showTicks =FALSE
, groupnameFontsize = 8 # have to shrink font for web viewing
, groupnamePadding = 5
, groupThickness = .05
, chordedgeColor = "gray90"
, groupColors = getPalette(colorCount)
)
In the interactive graph below, the diagonals have been removed to give space to emphasize the connnections. But the code-chunk above will produce one with the diagonals still in.
# without the diags means there's NOT return
chorddiag::chorddiag(remove_diags(adjmat[ord, ord], FALSE), margin = 150, showTicks =FALSE
, groupnameFontsize = 8
, groupnamePadding = 5
, groupThickness = .05
, chordedgeColor = "gray90"
, groupColors =getPalette(colorCount))
If you’re using RStudio, this figure should show in your Viewer. You can download it by selecting the Export tab, and choosing html option.

